Introduction
Our lab focuses on developing solutions to combat antimicrobial resistant bacteria and other emerging pathogens from various angles. We use an evolutionary-informed approach by understanding bacterial adaptation and leveraging microbial evolution to develop new treatments and detection methods.
#1
1- In terms of engineering controls for mitigation, treatment, and prevention of infectious disease, we work to:
- Inform design of next-generation nano-enabled antimicrobial material
First numbered bullet
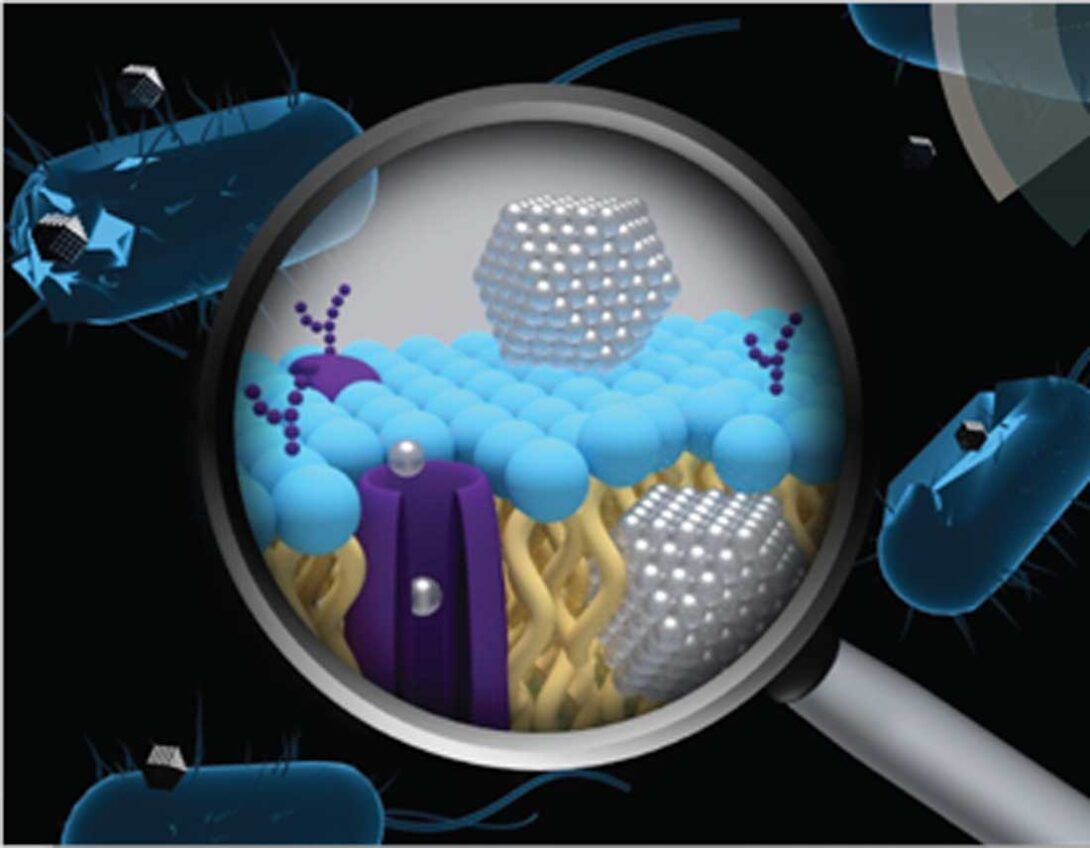
This project explores the interactions between engineered nanomaterials and bacteria, particularly for ionizing metal and metal-oxide nanoparticles, including:
- Evaluating particle-specific and ion-driven mechanisms of antimicrobial activity
- Establishing relationships between NP properties and antimicrobial activity
- Studying the microbial adaptive response to nanomaterial stress and understanding evolutionary mechanisms for adaptive bacterial resistance towards nanomaterials
- Advancing clinical application of nano-enabled antimicrobial materials
- Understand particle-specific and ion-driven mechanisms of action
Second bullet
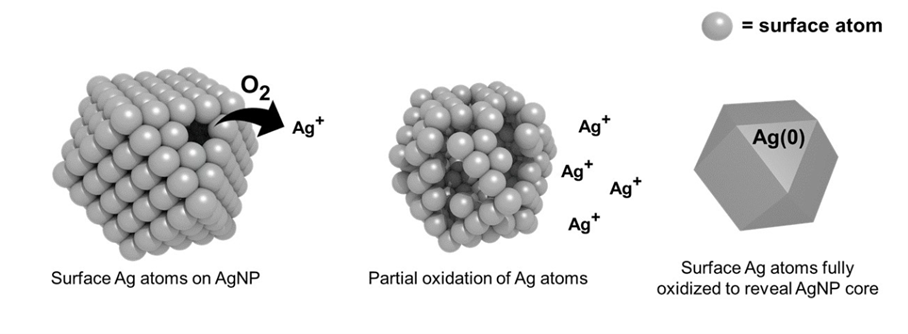
- Understand how we can harness microbial evolution and fitness as strategies for novel antimicrobial design
2
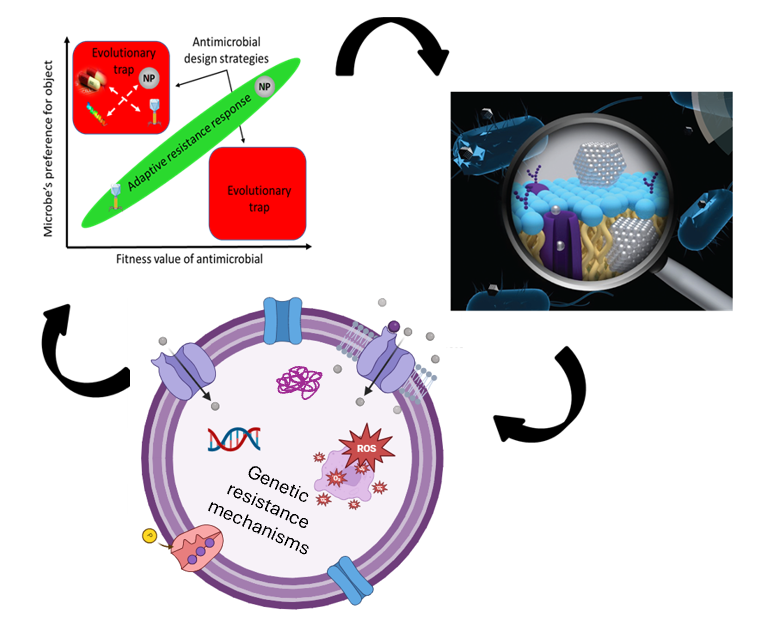
2- Pathogen detection is a first critical step in disease prevention and control. Thus, our lab aims to develop novel methods for rapid detection of resistant bacteria and broader pathogen detection that could be used for both clinical and wastewater surveillance/environmental monitoring. For this, we are exploring non-canonical genotype-to-phenotype relationships to establish new physiologically-based measurement capabilities.
3
3- Understanding the basis for and different factors influencing the evolution of resistant bacteria in the environment as well as the environmental impacts arising from AMR.
- Understand propagation of resistance in environmental microbial communities
- Identify new emerging resistances under changing environmental conditions
- Assess the environmental impacts on the evolution of antibiotic resistance
- Develop new monitoring targets for AMR
- Characterize changes in environmental microbiomes and functions due to AMR
We utilize the following methods and experimental workflows in our lab:
1- Nanoparticle synthesis and characterization – tuning nanomaterial properties as design handles
2. Classic laboratory evolution in single strain and mixed microbial systems
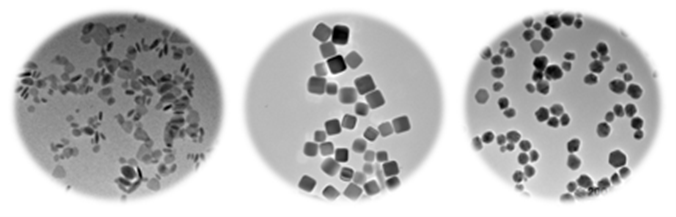
2- Classic laboratory evolution in single strain and mixed microbial systems
3-Advanced molecular
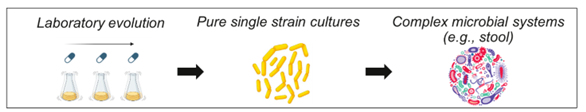
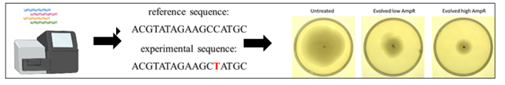
3- Advanced molecular characterization techniques (e.g., next-generation sequencing) and traditional, culture-based microbiological methods
3. Advanced
.